Research
Research Overview–
We are studying the molecular basis of translational control of the Hepatitis C Virus and Coxsackievirus B3 RNAs. Translation of these viral RNAs are guided by Internal Ribosome Entry Site (IRES). Ribosome binding to the IRES element is mediated by a number of cellular proteins (trans-acting factors) and cis acting RNA elements. Such RNA-protein interactions that are critical for viral IRES function, form a major research area in the lab. In this context, the group has expanded its scope to investigate the mechanistic details of HCV and CVB3 replication, host-viral interactions and related aspects of immunity. Recently, studies have been extended to study the role of the cellular host factors viz. proteins, long non-coding RNAs and micro RNAs in virus induced cellular pathogenesis. In addition, our group has extended study to non-canonical modes of cellular RNA translation using tumor suppressor p53 RNA as a model system.
HEPATITIS C VIRUS :
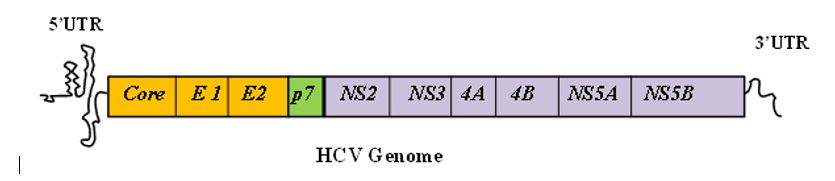
Hepatitis C virus (HCV), causing viral hepatitis, belongs to Hepacivirus genus of the Flaviviridae family. The error prone viral genome replication enables host immune evasion, which in turn facilitate viral persistence, leading to liver fibrosis, cirrhosis and hepatocellular carcinoma. HCV consists of a 9.6-kb single-stranded positive-sense RNA genome containing an open reading frame (ORF) flanked at both ends by highly structured and conserved untranslated regions (5’ and 3’UTRs). HCV translation is mediated by an internal ribosome entry site (IRES) located within the 341-nucleotide (nt) of 5′ UTR which further extends 30 to 40 nt downstream of the initiator AUG codon. On the other hand, HCV replication is mostly mediated by the 3′ UTR, which synthesizes a negative strand RNA. This negative-strand RNA then serves as a template for the generation of the plus-strand copies of viral RNA.
The error prone replication for HCV also leads to a challenge for development of specific antiviral therapies against all viral genotypes. The current therapy includes a combination of pegylated-IFNα (peg-IFNα) and ribavirin with either telaprevir or boceprevir. However, this triple therapy regimen has additional limitations due to selection of resistance variants.
The major focus of the study in our lab is to understand the basics of the various biological processes of the virus and implementation of the knowledge for the development of effective and suitable antivirals targeting those processes by use of different high-end technologies.
TRANSLATION
Several cellular trans-acting factors are known to bind to HCV IRES and influence the internal initiation. Our lab has characterized interaction of La (a cellular trans-acting factor), with HCV IRES, both in vivo and in vitro. (Pudi et al. Journal of Biological Chemistry, 2003 and 2004). We have shown that among the three RRMs of La protein, the RRM2 interacts with HCV internal ribosome entry site (IRES) around the GCAC motif near the initiator AUG present in the stem region of stem-loop IV (SL IV).
We have further demonstrated that La protein binding near the initiator AUG facilitates the interactions with ribosomal protein S5 and 48 S ribosomal assembly and influences the formation of functional initiation complex on the HCV IRES RNA to mediate efficient internal initiation of translation (Pudi et al. Journal of Biological Chemistry, 2004).
REPLICATION
Different stages of hepatitis C virus (HCV) life cycle are coordinated by a complex interplay of cis-acting elements, viral proteins and host factors. One of these stages is replication. HCV non-structural protein 3 (NS3) plays crucial role in viral polyprotein processing and RNA replication whereas human La protein (a cellular factor) is important for both HCV translation as well as replication. We have shown thatthe competition between the host factor (La) and the viral protein (NS3-protease) for binding to HCV-IRES contribute to the molecular switch from translation to replication of the HCV-RNA (Ray and Das, Scientific Report, 2011).
We have also demonstrated the mechanism of regulation of HCV replication by interaction of cis-acting element GCAC within HCV IRES with human La protein, unravelling yet one more specific target to inhibit HCV replication (Kumar et al, Journal of Virology, 2013).
In collaboration with Prof. Siddhartha Roy, IICB, Kolkata, we have shown that a highly conserved unique beta-turn in human La protein may contribute to host tropism of HCV (Kumar and Manna et al, Journal of Virology, 2014).
Following this, we explored other cellular IRES trans-acting factors (ITAFs) for their regulation of HCV replication. We have worked on the RNA binding proteins HuR and PTB. We show that HuR displaces PTB which prevents replication initiation at viral 3’UTR and assists in binding of La protein to the HCV 3’UTR, which is followed by replication complex assembly. (Shwetha et al, Journal of Virology, 2015). These ITAFs are nuclear resident proteins but get relocalised to cytoplasm upon viral infection exerting their role in viral lifecycle. Currently, our lab is interested in exploring the mechanism of this relocalisation and the cellular effects of the relocalisation.
We have further shown that HuR is regulated by miR125b-5p which gets upregulated during HCV infection, and targets HuR. We propose
this as a host response to virus induced relocalisation of HuR to the cytoplasm. To combat the increased availability of HuR in the cytoplasm, the host upregulates a microRNA which targets HuR, keeping a check on viral replication (Shwetha et al., Virus Research, 2018). Our research work would further provide detailed insights for better understanding of such a complex process.
We are further studying some more lncRNAs and miRNAs which affect viral lifecycle, directly or indirectly and the mechanism of their regulation under the condition of HCV infection.

NON-CODING RNA
Along with the host proteins, the HCV viral life cycle is also regulated by non-coding RNAs. The primary infection site of the virus is the liver. Highly Upregulated in Liver Carcinoma (HULC) is the lncRNA which is abundantly and uniquely expressed in Liver. We have studied its role in viral life cycle and found that it assists in viral release by regulating association of HCV core protein to the lipid droplets, which serve as a means of viral release (Sharma et al., Cellular Microbiology, 2019). Further we have also shown that HULC exerts a direct influence on viral replication by sponging an essential microRNA, miR122. We show that the sponging of miR122 by HULC prevents it from displacing PCBP2 from HCV 5’UTR, thereby having an inhibitory effect on viral replication (Sharma et al., Manuscript under preparation). We have also shown that exosome-associated microRNA-375 induces cell proliferation by regulating IGFBP4 upon hepatitis C virus infection (Pradhan et al. Molecular Microbiology, 2022).
ANTIVIRALS
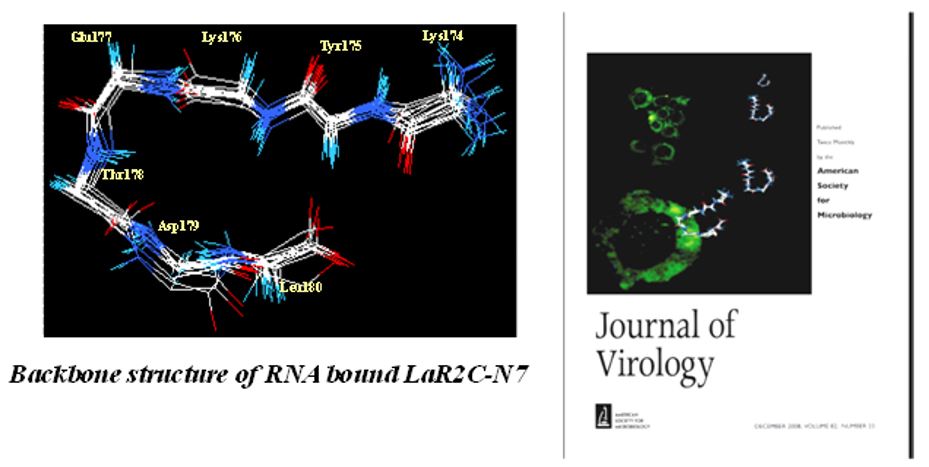
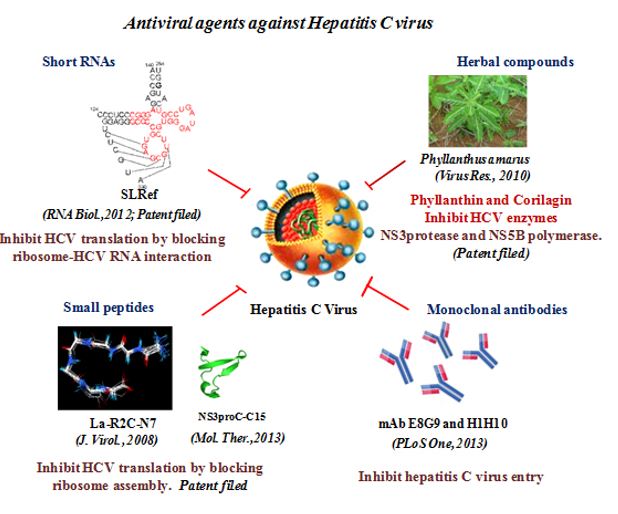
HCV-induced hepatitis is a serious medical problem of modern age with more than 170 million chronic carriers throughout the world. Chronic HCV infection can lead to liver cirrhosis and hepatocellular carcinoma (HCC). No vaccine is available yet. Different anti-HCV agents have been explored in recent past. Our research group demonstrated that a synthetic peptide LaR2C, derived from La-RRM (112-184) could interact with HCV RNA and interferes with the formation of 48S initiation complexes by acting as a dominant negative inhibitor of viral RNA translation (Pudi et al, Journal of Virology, 2005). Further, using NMR spectroscopy of the HCV IRES bound La-peptide (in collaboration with Dr. Siddhartha Roy, IICB) we have demonstrated that the residues responsible for RNA recognition form a turn in the RRM structure. More importantly, a hepta-peptide comprising this turn showed significant inhibition of replication of HCV monocistronic replicon in Huh7 cells (Mondal and Ray et al, Journal of Virology, 2008).
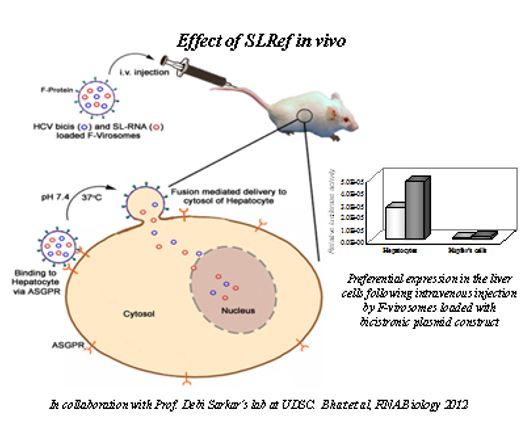 We have targeted the translation-replication switch using a peptide derived from the RNA binding region of the NS3pro protein. A 30-mer peptide was designed from the predicted RNA-binding region of NS3 protease that inhibited IRES-mediated translation. Further, this peptide was truncated to 15-mer and this could also inhibit HCV translation as well as replication (in collaboration with Prof. N. Srinivasan, MBU, IISc). More importantly, in collaboration with Prof. Debi Sarkar, UDSC, we tested its activity in an in vivo mouse model using Sendai virus based virosome system (Ray and Roy et al, Molecular therapy, 2013).
We have targeted the translation-replication switch using a peptide derived from the RNA binding region of the NS3pro protein. A 30-mer peptide was designed from the predicted RNA-binding region of NS3 protease that inhibited IRES-mediated translation. Further, this peptide was truncated to 15-mer and this could also inhibit HCV translation as well as replication (in collaboration with Prof. N. Srinivasan, MBU, IISc). More importantly, in collaboration with Prof. Debi Sarkar, UDSC, we tested its activity in an in vivo mouse model using Sendai virus based virosome system (Ray and Roy et al, Molecular therapy, 2013).
Additionally, our group (in collaboration with Dr. Akhil Banerjea, NII) demonstrated an efficient use of DNAzymes as inhibitor of HCV-RNA translation and replication (Roy et al, Journal of General Virology, 2008). Our laboratory (in collaboration with Prof. Debi Sarkar, UDSC) successfully demonstrated efficient delivery of antiviral shRNAs into mouse liver using Sendai virus fusion protein-based virosome system, and inhibition of HCV-RNA translation in whole animal (Subramanian et al. Journal of General Virology,2009). We also employed a novel approach to inhibit viral RNA translation using small RNA molecules derived from structured domains of HCV-IRES element. A small RNA molecule analogous to the SL III e+f subdomain of the HCV IRES inhibited translation very efficiently and selectively. This RNA interacts with the 40S ribosomal protein S5 and blocks the interaction between the ribosome and HCV IRES (Ray et al, Nucleic Acids Research, 2004).
More importantly, using Sendai virosome we have preferentially delivered this small RNA into mice liver and demonstrated inhibition inhibit HCV RNA (Bhat et al., RNA Biology, 2012). These observations constitute the “proof of the concept” that the ‘ribosome-viral RNA’ interaction can be specifically targeted to develop novel antiviral to combat HCV infection.
Antivirals targeting HCV enzymes: Herbals
In view of safe and effective drug development for anti HCV therapy, we identified, Phyllanthus amarus (Euphorbiaceae), a natural herbal plant, which inhibits HCV viral enzymes and replication most significantly (Ravikumar et al, Journal of virus Research,2011).
We have identified a principal bioactive component ‘corilagin’ from Phyllanthus amarus. This pure compound could effectively inhibit viral replication in the infectious cell culture system, displayed strong antioxidant activity by blocking HCV induced generation of reactive oxygen species and suppressed up-regulation of NOX4 and TGF-β mRNA levels (Reddy et al., Antiviral Research, 2018). We have also identified certain chemical compounds from the crude extract of the fruit Pomegranate (Punica granatum). The pure compounds, punicalagin, punicalin, and ellagic acid isolated from the extract specifically blocked the HCV NS3/4A protease activity in vitro (Reddy et al., Scientific Reports, 2014). Our study provides a proof-of-concept approach for the potential use of antiviral and non-toxic principle ellagitannins from pomegranate in prevention and control of HCV induced complications.
VACCINE
Our lab (as part of an Indo-Australian project) has also initiated work on HCV vaccine inducing both humoral and cell mediated immunity. We are trying to develop a vaccination regimen to generate antibody-based and cell mediated immunity against hepatitis C virus against multiple HCV genotypes. This is being done by simultaneous vaccination with virus-like particles (VLPs) containing the core and envelope proteins for neutralizing antibody generation and DNA vaccine construct encoding selective non-structural proteins for inducing cell mediated immunity. The results of vaccination with the individual components of the vaccine are being measured in first mice model and then pig model. These studies are expected to yield an effective HCV vaccine able to generate effective immunity against multiple genotypes in humans.
PATHOGENESIS
The current focus of our lab also involves the study of miRNA and proteome profiles in the serum of HCV infected patients. We have carried out a 2-DE based proteomic analysis and identified differentially regulated proteins in serum of Indian patients infected with HCV genotypes 1 and 3.
We have also used JFH1 infectious cell culture system to confirm our observation and have characterized the role of RBP4 in HCV infection in detail. (Gouthamchandra et al, Journal of General Virology, 2014).
In parallel, we have performed miRNA microarray studies with RNA from HCV infected serum samples. We have done the serum miRNA profiling of patients harbouring HCV genotype 1 or genotype 3 virus (Shwetha et al., Scientific Reports, 2013) to study the deregulated miRNAs.
Further, extracellular miRNAs in the serum are protected from degradation by either being associated with RNA binding proteins or by being incorporated in the vesicles such as exosomes. Exosomes are reported to carry certain microRNAs as well as HCV genomic RNA. We have studied the deregulated miRNAs and proteins in exosomes isolated from HCV infected individuals as well as from cell culture supernatant. We are currently characterising these miRNAs in HCV life cycle and pathogenesis.
MECHANOBIOLOGY OF LIVER CELLS
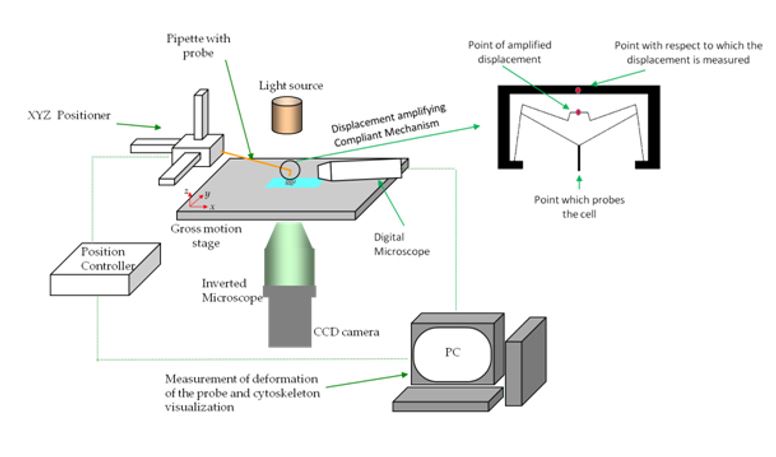
In collaboration with Prof. Ananthasuresh from the Mechanical Engineering department, IISc, we are studying the response of liver cells to mechanical stimuli for identification of mechanical biomarkers for disease states specifically hepatitis and cancerWe intend to look at the effects at single cell level and at population level.
For looking at response at the single cell level, we are developing various compliant mechanisms for applying required forces on to cells and imaging fluorescently labelled proteins for eliciting the response.
At the population level, we are looking at the response of cells to flow stimuli. For inducing the cells to flow stimuli we have developed convenient perfusion culture systems consisting of miniature bioreactors, miniature perfusion pumps and electronic controls (Sreenath et al., Biores Open Research, 2015) . We are interested in the differential regulation of genes and proteins specifically those concerning the mechanical properties of the cell like the cytoskeleton and the cell membrane. We have shown that HCV alters the morphology and the stiffness of the nuclei by down-regulating lamin A/C, rendering the nuclear morphology susceptible to shear stress (Sreenath et al., Biophysical Journal, 2019). By subjecting the cells to shear stress due to flow, we show that the nuclei of HCV infected cells assume increasingly elliptical shape even at low shear stress corresponding to interstitial flow. We believe that HCV is exploiting the deregulation of nuclear lamina for its pathogenesis.
Coxsackievirus B3
Coxsackievirus B3 (CVB3) is an Enterovirus, known to infect humans causing myocarditis and less commonly, pancreatitis and aseptic meningitis. Its genome is a positive sense RNA. The genome is 7.5 kb long consisting of coding region flanked with 5’ and 3’ untranslated region (UTR).
CVB3 Genome
CVB3 consists of a type I IRES similar to polio virus. These IRES require both canonical translation initiation factors and IRES trans acting factors (ITAFs) derived from the host for translation initiation. Upon entry in the host cell, viral proteins are synthesized. The virus is known to hijack the host translational machinery. It shuts off the cap-dependent translation and utilize the host factors for the viral translation and replication.
CVB3 UTRs in viral life cycle
It is well established for the viral UTRs to be highly structured and divided into domains that play specific role in viral translation and replication. We have studied the importance of some conserved sequences and structures in the CVB3 5’ UTR such as the GAGA loop, spacer region, SL C/c apical loop and cryptic AUG as integral factors for viral translation. These cis acting factors are important for their ability to interact with cellular and viral proteins. (Bhattacharyya et al, Virus Research, 2004; Bhattacharyya et al, RNA Biology, 2006; Bhattacharyya and Verma et al, Virology, 2008) Our work indicates that mutating the cryptic AUG residue leads to inhibition of interaction of some host factors with viral RNA as well as decreased 48S ribosomal complex interaction and hence affects viral translation. (Verma et al, Journal of General Virology, 2011).
Host factors involved in viral life cycle
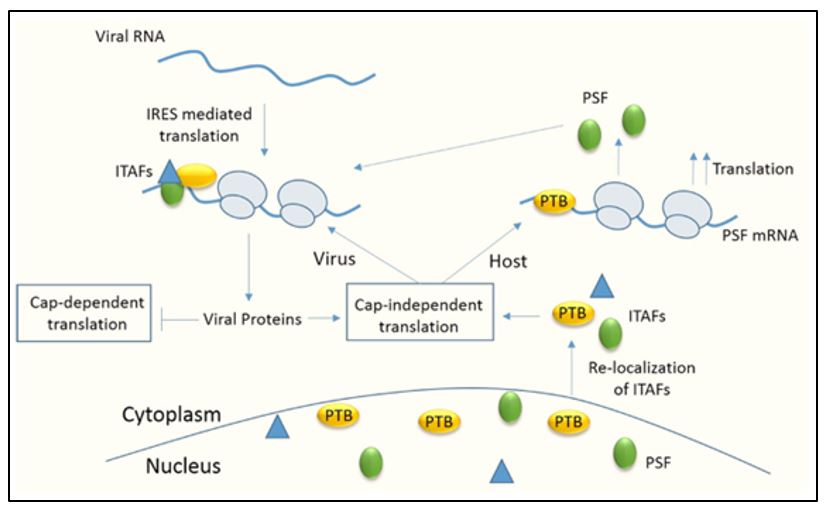
As mentioned earlier, initiation through the viral IRES requires binding of various host factors. We have tried to study the role of various host factors interacting with the IRES. We have identified La (Ray et al, Nucleic Acids Research, 2002), Poly-pyrimidine tract binding protein (PTB) (Verma et al, Journal of General Virology, 2010) and PTB associated splicing factor (PSF) (Dave et al, Nucleic Acids Research, 2017), as ITAFs required for viral IRES dependent translation. Knockdown of these proteins specifically inhibits viral translation. We also found that there is relocalization of certain host factors during infection from nucleus to cytoplasm to aid viral life cycle.
Role of PSF during CVB3 infection
C-terminal fragment of eIF4GI, characterized for its role in viral translation, is generated after cleavage mediated by viral protease 2A. However, 2A protease is not present in the capsid and is synthesized only after the initial round of translation. This presents the conundrum of how the initial round of translation occurs in the absence of the C-terminal eIF4GI. We have characterized an isoform of eIF4GI, known as Death Associated Protein 5 (DAP5), for its role in initial round of translation. Because of its lower affinity for the viral IRES as compared to C-terminal eIF4GI, DAP5 is later replaced by cleaved eIF4GI for robust viral RNA translation. (Dave et al., Journal of Biological Chemistry, 2019)
After viral translation, the positive sense RNA undergoes switch from translation to replication. We have identified a host protein, hnRNP C1/C2 which guides this switch. It interacts with stem-loop V in the IRES and displaces PTB, a positive regulator of translation. (Dave et al., RNA Biology, 2019)
Further, we are studying the involvement of host factors in CVB3 replication. We have identified HuR as a negative regulator of viral replication. It gets upregulated upon CVB3 infection and this increase is mediated by downregulation of a particular microRNA, miR125b, through viral 2A protease. (Biju et al., RNA Biology, 2021) We are currently interested in investigating the role of HuR in pathogenesis of the virus.
SARS-CoV2-
Covid-19 is an upper respiratory tract infectious disease caused by SARS-CoV-2, an RNA virus belonging to the Coronaviridae family. Being a novel corona virus, the specific therapeutics has not been developed yet. Hence, a specific vaccine for the SARS-CoV-2 is urgently needed to control the pandemic. SARS-CoV-2 virus infects the cells through the ACE2 receptor, which is recognized specifically by the Spike protein, one of the structural proteins of SARS-CoV-2 virus. Studies have shown that these structural proteins elicit host Humoral immune response. Therefore, many research groups have implemented such mechanisms to generate vaccine against viruses. Hence, we are working to make Virus Like Particles (VLP) for the development of vaccine. VLP is composed of only the structural proteins that are non-infectious but generate immune response similar to infectious virus particles. Using this VLP approach, we are also studying the immunogenic implications of the different mutations (Raheja et al. Microbiology Spectrum, 2022).
p53
Translational regulation of p53 mRNA by trans-acting factors and cis-acting elements
Tumor suppressor protein p53 is popularly considered as “guardian of the genome”. It is a master transcription regulator that is indispensable for controlling several cellular pathways. Its prime role is to adapt gene expression programs in order to maintain cellular homeostasis and genome integrity especially during the stress conditions. Most, if not all, tumors harbor mutations in the TP53 gene itself or acquire genetic and/or epigenetic alterations that compromise the p53 response. The regulation of p53 expression and activity is multi-layered. It is intricately controlled at the levels of transcription, RNA splicing, RNA stability, post-transcriptional gene-silencing, translation, post-translational modifications, protein stability and degradation. Extensive research has been carried out on regulation of p53 activation and function at the protein level, but translation of p53 mRNA is the focus of current research activities as this is one of the few steps in the regulation of any gene where response to any stimulus can be swiftly graded up or down in terms of the protein output.
Work done in our laboratory demonstrated that p53 mRNA has a dual Internal Ribosome Entry Site (IRES) structure that regulates translation of full-length p53 and its N-terminally truncated isoform ΔN-p53. The two IRESs show distinct cell-cycle phase-dependent activity with the IRES for full-length p53 being active at the G2–M transition and the IRES for ΔN-p53 shows highest activity at the G1–S transition (Ray et al, EMBO Reports, 2006). Translation from p53 IRESs depend upon IRES trans-acting factors (ITAFs) as well as cis-acting structural elements. We have demonstrated stress-dependent modulation of p53 IRESs by ITAFs. Polypyrimidine tract-binding protein (PTB) binds specifically to both the p53 IRESs but with differential affinity. During doxorubicin treatment, PTB protein translocates from nucleus to the cytoplasm, suggesting that the relative cytoplasmic abundance of PTB protein, under DNA-damaging conditions, might contribute to regulating the coordinated expression of the p53 isoforms, owing to the differential affinity of PTB binding to the two p53 IRESs (Grover et al, Cell Cycle. 2008). Further, using RNA affinity approach we have identified Annexin A2 and PTB associated Splicing Factor (PSF/SFPQ) as p53 ITAFs. Interestingly, in the presence of calcium ions Annexin A2 showed increased binding with p53 IRES. More importantly the interplay between Annexin A2, PSF and PTB proteins for binding to p53 mRNA appears to play a crucial role in IRES function (Sharathchandra et al, RNA Biology, 2012). In a collaborative project with Prof Adi Kimchi’s lab (Weizmann Institute of Science) we also found that an auxiliary translation initiation factor, eIF4G homolog Death Associated Protein promotes IRES-driven translation of p53 mRNA. Functional analysis indicated that DAP5 preferentially promotes translation from the second module of IRES residing in the coding sequence (Weingarten-Gabbay S and Khan et al., Oncogene, 2014).
To correlate p53 IRES structure with activity, we have mapped the putative secondary structure of p53-IRES RNA using information from chemical probing and nuclease mapping experiments. Compared to wild-type RNA, our results also indicate weaker and compromised IRES activities of the p53 IRESs bearing cancer-derived silent mutations in the 5’UTR or in the coding region of the IRES. Further, it was observed that few cytoplasmic trans-acting factors such as PTB and hnRNP C1/C2, critical for enhancing IRES function, were unable to bind these mutant IRESs as efficiently as in wild-type. In all these cases, knockdown of ITAFs or mutations in p53 IRESs show consequent changes in p53 dependent transactivation of target genes (Sharathchandra et al, 2012, RNA Biology; Khan et al., Oncogene, 2012). Thus IRES-dependent translational control of p53 mRNA is uniquely indispensable in the complex scheme of p53 isoforms activation and function.
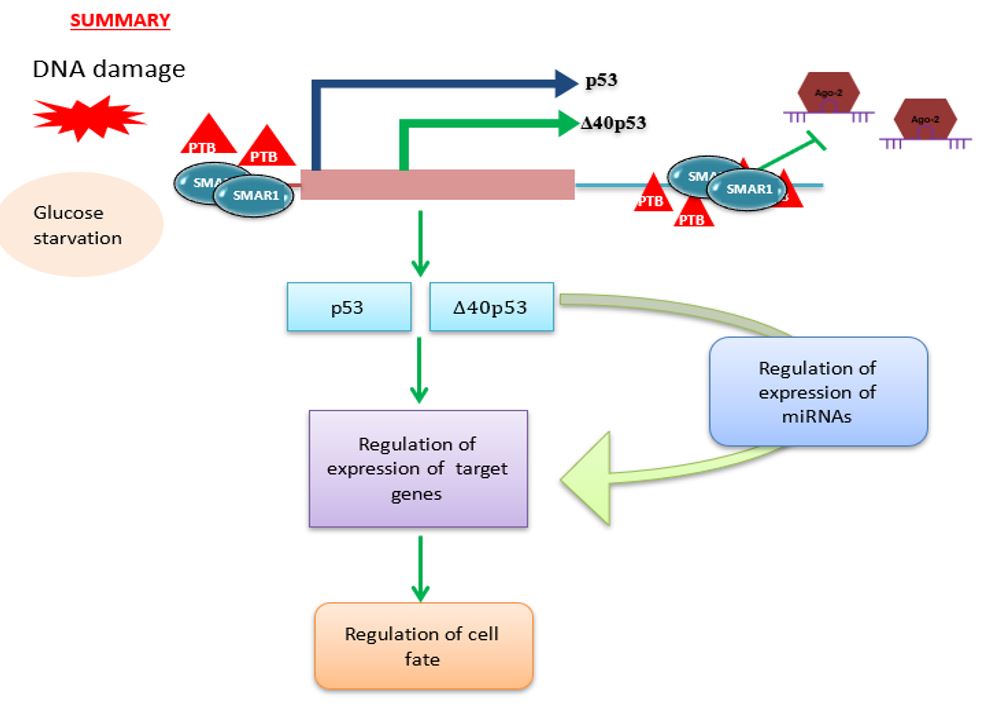
Deregulations in translational control contribute to each step of cellular transformation and tumor progression; nutrient starvation being a very common stress for the cells located inside non-vascularized tumors. Therefore, we have addressed nutrient-mediated translational regulation of p53 mRNA using glucose depletion. We found scaffold/matrix attachment region-binding protein 1 (SMAR1), a predominantly nuclear protein to be abundant in the cytoplasm under glucose deprivation. Importantly under these conditions polypyrimidine-tractbinding protein, an established p53 ITAF did not show nuclear-cytoplasmic relocalization highlighting the novelty of SMAR1-mediated control in stress. In vivo studies in mice revealed starvation-induced increase in SMAR1, p53 and Δ40p53 levels that was reversible on dietary replenishment. SMAR1 associated with p53 IRES sequences ex vivo, with an increase in interaction on glucose starvation. RNAi-mediated-transient SMAR1 knockdown decreased p53 IRES activities in normal conditions and under glucose deprivation, this being reflected in changes in mRNAs in the p53 and Δ40p53 target genes involved in cell-cycle arrest, metabolism and apoptosis such as p21, TIGAR and Bax. Activation of p53 IRESs on glucose deprivation provides a new regulatory aspect to the field of translational control of p53 mRNA, suggesting critical function of the p53 isoforms in these conditions in terms of cell survival and stress tolerance (Khan et al, Cell Death and Differentiation, 2015).
We have also investigated the effect of p53 ITAFs in regulating cross-talk between 5’ UTR and 3’UTR of p53 mRNA and the role of miRNA and protein interactions at the 3’UTR in regulating p53 and ΔN-p53 expression from p53 mRNA. We have reported that there is an interplay between miRNAs and PTB at the p53 3’UTR under normal and stress conditions like DNA damage. Interestingly, PTB showed some overlapping binding regions in the p53 3’UTR with miR-1285. In fact, knockdown of miR-1285 as well as expression of p53 3’UTR with mutated miR-1285 binding sites resulted in enhanced association of PTB with the 3’UTR, which provides mechanistic insights of this interplay. Taken together, the results provide a plausible molecular basis of how the interplay between miRNAs and the PTB protein at the 3’UTR can play pivotal role in fine tuning the expression of the two p53 isoforms (Katoch et al, Nucleic Acids Research, 2017).
Recently we have been also studying how p53 isoforms regulate the expression of cellular coding as well as non-coding RNAs. Results from the miRNA microarray showed that there are different miRNAs whose expressions are regulated under Δ40p53 expression. These miRNAs were found to be involved in different pathways namely, cell cycle regulation, apoptosis, cell proliferation, senescence etc. The results unravel a new dimension to our understanding of Δ40p53 functions, which can also regulate cellular fate independent of p53FL (Katoch et al., Cell Cycle, 2021). We are also currently studying the role of isoforms in regulating the expression of long non-coding RNAs.
Thus the overarching aim of p53 project is to identify novel factors which regulates p53 isoforms and also factors which been regulated by p53 and Δ40p53.
Group Member

Harsha Raheja
Graduate Students
Year of Joining: 2015 (Int. PhD)
B.Sc. (Hons) Microbiology- University of Delhi
E-mail- harsharaheja@iisc.ac.in
Title: Studies on host-virus interaction in HCV infection

Sachin Kumar Tripathi
Graduate Student
Year of Joining: 2016
M.Sc. (Biotechnology): TMBU, Bhagalpur (Bihar)
E-mail- sachint@iisc.ac.in
Title: HCV induced HCC: Role of lncRNAs

Apala Pal
Graduate Students
Year of Joining – 2017
M.Sc. (Biochemistry): University of Calcutta
E-mail- apalapal@iisc.ac.in
Title: Understanding the role of p53 translational isoforms in regulating cellular fate

Priya Rani
Graduate Students
Year of Joining- 2017
M.Sc. (Biotechnology): University of Hyderabad
E-mail- priyarani@iisc.ac.in
Title: Role of host factors in CVB3 life cycle and pathogenesis

Ashish Aneja
Graduate Students
Year of Joining- 2017 (Int. PhD)
B.Sc. (Hons) Botany- University of Delhi
E-mail- ashishaneja@iisc.ac.in
Title: Role of cis elements and trans factors in Dengue life cycle and pathogenesis

Anwesha Barua
Graduate Students
Year of Joining- 2018
B.Tech- VIT
E-mail- anweshabarua1996@gmail.com
Title: Checking the mechanical and biological response ofliver cells on gels of different stiffness
Risabh Sahu
Graduate Students
Year of Joining- 2020 (Int. PhD)
B.Sc. (Hons) Microbiology- Ramakrishna mission Vidyamandira(Autonomous college)
E-mail- risabhsahu@iisc.ac.
Title: Dengue viral infection, pathogenesis and immunology
Pritam Kumar Ghosh
Graduate Students
Year of Joining- 2020 (Int. PhD)
B.Sc. (Hons) Life Sciences- Presidency University, Kolkata
E-mail- pritamkumar@iisc.ac.in
Title: p53 and cancer biology

Dr.Ashutosh Shukla
Postdoctoral Fellow
Ph.D. Indian Institute of Technology, Delhi
Email- ashutoshshuklaiitd@gmail.com
Awards & Patents
Patents from IISc
- * ‘A Small Synthetic RNA, A Method of preparing the same and uses there of’–by ParthoSarothi Ray and Saumitra Das. (Application no. 224/CHE/2004, dated 12.03.2004; published in the Patent office Journal, dated 13.01,06. (WO/2005/087923) APPROVED
- * ‘Antiviral Peptide against Hepatitis C virus’ by RenukaPudi, Sudhamoni Sonny and Saumitra Das (Application no. 520/CHE/2005, dated 02.05.2005. PCT filed (WO/2006/117805) APPROVED
- * Small antiviral Peptides AGAINST Hepatitis C Virus:Saumitra Das, Siddhartha Roy, Upasana Ray, TanmoyMondal, Asit Kumar Manna, Romi Gupta. filed May 2, 2005.
- ‘Compositions method and formulations for peptide based inhibitors of hepatitis C virus replication’by Saumitra Das et al, Patent filed 4033/CHE/2011, 23/11/2011
- ‘Phyllanthin as a potent inhibitor of Hepatitis C virus replication’, by Saumitra Das et al,4034/CHE/2011, patent filed 23/11/2011
Patents from UCLA
- +Selective Inhibition of Internally Initiated RNA translation. By Saumitra Das, Asim Dasgupta and Peter Coward. US Patent application No. 08/817, 953
- +Interference with viral IRES-mediated translation by a small yeast RNA reveals critical RNA-Protein interactions. By Saumitra Das and Asim Dasgupta. US Patent application no. 09/316,630
- +Methods to inhibit viral replication. By Asim Dasgupta, Saumitra Das and Narayan Baidya. US patent application no. 09/836,073
Alumni
Graduate Students
| Name | Year of Graduation | Current Position |
| Renuka Pudi | 2004 | Lead Research Scientist at LDRTC Washington D.C. Metro Area |
| Partho Sarothi Ray | 2004 | Associate Professor at IISER, Kolkata |
| Shankar Bhattacharya | 2006 | Research Scientist D at Translational Health Science and Technology Institute, NCR, Delhi |
| Debojyoti Dhar | 2008 | Founding Member & Director (Business Development and Innovation) at Leucine Rich Bio Pvt Ltd, India |
| Richa Grover | 2009 | Faculty Scientist at Institute of Bioinformatics & Applied Biotechnology, Bangalore |
| Bhupendra Kumar Verma | 2010 | Assistant Professor at AIIMS, New Delhi |
| Upasana Ray | 2011 | Assistant Professor at IICB, Kolkata |
| Sharath Chandra Arandkar | 2013 | Assistant Professor at ACTREC, Mumbai |
| Debjit Khan | 2014 | Postdoctoral Research Fellow at Cleveland Clinic, Ohio |
| Prasanna Bhat | 2014 | Postdoctoral Research Fellow at University of Texas Southwestern Medical Centre |
| Anuj Kumar | 2014 | Postdoctoral Researcher at Centre International de Rechereche en Infectiologie, France |
| S. Shwetha | 2015 | Postdoctoral Researcher at Stanford University, California |
| Aanchal Katoch | 2018 | Postdoctoral Researcher at Yale University, Connecticut |
| Pratik Dave | 2018 | Postdoctoral Researcher at FMI, Switzerland |
| Geetika Sharma | 2018 | Postdoctoral Researcher at The Rockefeller University, New York |
| Aunji Pradhan | 2021 | |
| Collaboration Students | ||
| Abhirami Lakshminarayanan (Org. Chem) | 2015 | Postdoctoral Researcher at University of Oxford, UK |
| Sreenath Balakrishnan (BSSE) | 2018 | Assistant Professor at IIT-Goa |
Postdoctoral Fellows
| Name | Tenure | Current Position |
| Dr. Nisha Rathore | 1999-2001 | Research at Genentech San Francisco Bay Area |
| Dr. Sudhamani SR | Postdoctoral Research Fellow at NIH | |
| Dr. M. Venkataramana | 2006-2009 | Associate Professor, University of Hyderabad |
| Dr. Ravi Kumar.Y.S | 2007-2010 | Associate Professor at M. S. Ramaiah Institute of Technology, Bengaluru |
| Dr. Raja Naika | 2009-2010 | Associate Professor, Tumkur University |
| Dr. Abraham Joseph | Associate Professor at University of Calicut | |
| Dr. Goutham Chandra | 2010-2016 | Research Scientist at Vidya Herbs Pvt. Ltd. Bangalore, India |
| Dr. Arunima S | 2013-2014 | Postdoctoral Research Fellow at IIT Bombay |
| Dr. Ranajoy Mullick | 2013-2017 | Scientific Officer at THSTI, NCR, Delhi |
| Dr. Papai Roy | 2015 | Scientific Officer at Shri Gangaram Hospital, New Delhi |
| Dr. Uma Reddy | 2010-2019 | Associate Professor at Vijaynagara University |
| Dr. Biju George | 2016-2022 | |
| Dr. Padmanava Behera | 2018-2021 | Postdoctoral Research Fellow, ILS Bhuvneshwar |
| Dr. Trinath Ghosh | 2018-2021 | Postdoctoral Research Fellow, NIBMG, Kalyani |
Project Assistants
| Name | Tenure | Current Position |
| Chaitrali Laha Roy | 2008-2010 | Postdoctoral Researcher at NCBS, Bangalore Ph.D from French Institute of Health and Medical Research, Paris |
| Nanditha M | 2009-2012 | Ph.D student at Medical University of Graz, Austria |
| Debashree Gowswami | 2010-2011 | Postdoctoral Researcher at Seattle Children’s Research Institute Completed Ph.D from Max Planck Institute for Biomedicine, Muenster, Germany |
| Ridhima Lal | 2010-2013 | Postdoctoral researcher at Cancer Research UK, Glasgow Completed Ph.D from Medical University of Graz, Austria |
| Divya K Sharma | 2011-2014 | Research engineer at Siemens, Bangalore |
| Priyanka Lahiri | 2013-2014 | Ph.D Student at Molecular Biophysics Department, IISc, Bangalore |
| Sreya Das | 2014 | Ph.D Student at IIT Bombay |
| Deeptha Vasudevan | 2014 | Ph.D Student at University of Utah |
| Nilanjan Mukherjee | 2014 | Life Science Research Professional 2 at Baxter Labs, Stanford University |
| Ashutosh | 2014 | Ph.D Student at JNCASR, Bangalore |
| Anirban Mukherjee | 2015 | Junior Scientist at National Institute of Biologicals, Noida, UP |
| Yoshika Janapala | 2016-2017 | Ph.D Student at Australia National University, Canberra |
| Priyanka Srinivasan | 2017-2018 | Ph.D Student at Australia National University, Canberra |
| Rachana Srinivasa | 2017-2018 | Pursuing M.S degree from University of Bonn, Germany |